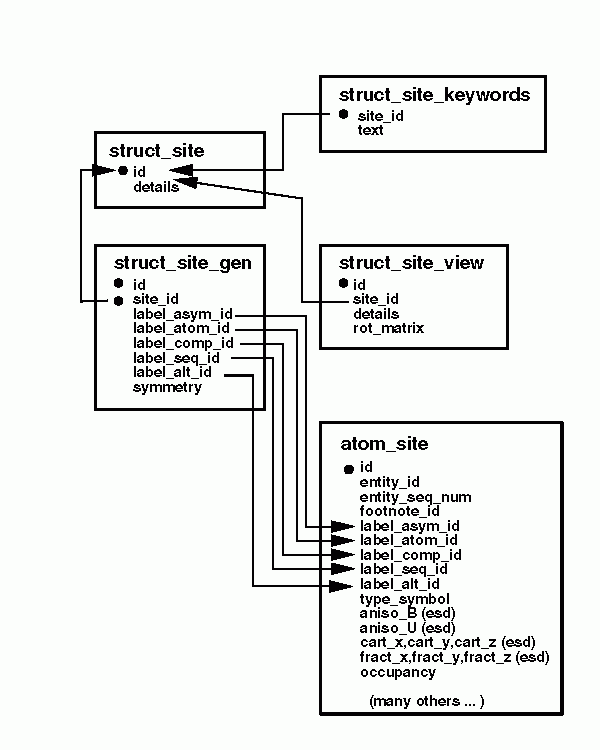
STRUCT_SITE. A textual description including
keywords and a series of views can be specified for each site.
Sites are generated from a list of structural features
and associated symmetry transformations. Structural features
forming a site are labeled using some combination of the identifiers
used to label atomic positions in the ATOM_SITE category.
These identifiers include the asymmetric unit identifier, component identifier,
sequence position, and the atom name. This is illustrated in the following diagram.
An example that describes the two binding sites in the HIV protease system (PDB 5HVP, Fitzgerald et al., 1990).
loop_ _struct_site.id _struct_site.details 'P2 site C' ; residues with a contact < 3.7 Angstroms to an atom in the P2 moiety of the inhibitor in the conformation with _struct_asym.id = C ; 'P2 site D' ; residues with a contact < 3.7 Angstroms to an atom in the P1 moiety of the inhibitor in the conformation with _struct_asym.id = D) ; # loop_ _struct_site_gen.id _struct_site_gen.site_id _struct_site_gen.label_comp_id _struct_site_gen.label_asym_id _struct_site_gen.label_seq_id _struct_site_gen.symmetry _struct_site_gen.details 1 'P2 site C' VAL A 32 1_555 . 2 'P2 site C' ILE A 47 1_555 . 3 'P2 site C' VAL A 82 1_555 . 4 'P2 site C' ILE A 84 1_555 . 5 'P2 site D' VAL B 232 1_555 . 6 'P2 site D' ILE B 247 1_555 . 7 'P2 site D' VAL B 282 1_555 . 8 'P2 site D' ILE B 284 1_555 .
An abbreviated example that describes the binding site for an intercalating drug in a drug-DNA complex (NDB DDF040, Leonard et al., 1993).
loop_
_struct_site.id
_struct_site.details
B1 'Binding at TG/AC Step 1'
loop_
_struct_site_keywords.site_id
_struct_site_keywords.text
B1 'Intercalation complex'
loop_
_struct_site_gen.id
_struct_site_gen.site_id
_struct_site_gen.label_asym_id
_struct_site_gen.label_comp_id
_struct_site_gen.label_seq_id
_struct_site_gen.symmetry
1 B1 A T 1 1_555
2 B1 A G 2 1_555
3 B1 A C 5 8_555
4 B1 A A 6 8_555
5 B1 D DM2 . 8_555
loop_
_struct_site_view.id
_struct_site_view.site_id
_struct_site_view.details
_struct_site_view.rot_matrix[1][1]
_struct_site_view.rot_matrix[1][2]
...
_struct_site_view.rot_matrix[3][3]
View1 B1
'View along the base-pair plane'
0.133 0.922 ... -0.172
loop_
_atom_site.id
_atom_site.label_atom_id
_atom_site.label_comp_id
_atom_site.label_asym_id
_atom_site.auth_seq_id
_atom_site.cartn_x
_atom_site.cartn_y
_atom_site.cartn_z
_atom_site.occupancy
_atom_site.B_iso_or_equiv
_atom_site.label_entity_id
_atom_site.label_seq_id
1 O5* T A 1 -18.744 20.195 22.722 1.00 36.68 1 1
18 P G A 2 -14.347 22.307 21.991 1.00 40.20 1 2
81 P C A 5 -3.756 13.529 9.644 1.00 31.11 1 5
106 O4* A A 6 -10.114 9.255 2.687 1.00 27.42 1 6
122 C1 DM2 D 7 -12.358 18.750 5.310 1.00 2.93 2 .
# ... Abbreviated ...
[an error occurred while processing this directive]