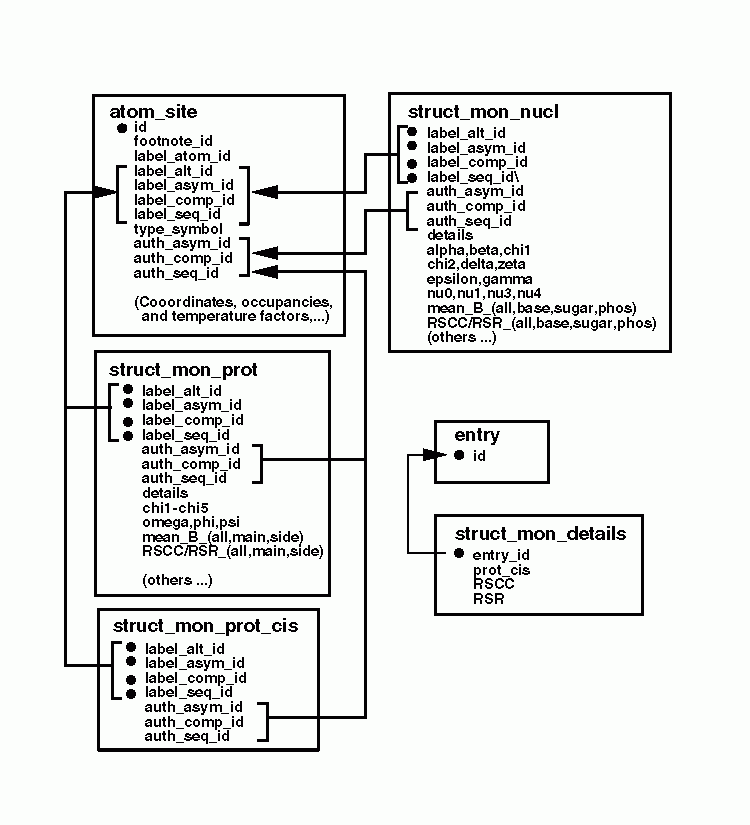
STRUCT_MON_DETAILS, STRUCT_MON_PROT,
STRUCT_MON_NUCL, and STRUCT_MON_PROT_CIS. These
categories contain features such as: backbone torsion angles, isotropic
temperature factors, real space R factors, and real space correlation coefficients.
Precise details about the calculation of the latter quantities may
be specified in the STRUCT_MON_DETAILS category. The
structural features described in these categories can be identified
using standard mmCIF monomer labels and/or an alternative nomenclature
preferred by the author. This is illustrated in the following diagram.
An abbreviated example showing showing the monomer properties for a single protein residue.
_struct_mon_prot.comp_id ARG _struct_mon_prot.seq_id 35 _struct_mon_prot.asym_id A _struct_mon_prot.alt_id . _struct_mon_prot.chi1 -67.9 _struct_mon_prot.chi2 -174.7 _struct_mon_prot.chi3 -67.7 _struct_mon_prot.chi4 -86.3 _struct_mon_prot.chi5 4.2 _struct_mon_prot.RSCC_all 0.90 _struct_mon_prot.RSR_all 0.18 _struct_mon_prot.mean_B_all 30.0 _struct_mon_prot.mean_B_main 25.0 _struct_mon_prot.mean_B_side 35.1 _struct_mon_prot.omega 180.1 _struct_mon_prot.phi -60.3 _struct_mon_prot.psi -46.0
This is another example which shows an abbreviated list of the nucleotide backbone conformation angles for a B-DNA duplex structure (NDB BDL028 Berman et al. 1991).
loop_ _struct_mon_nucl.label_comp_id _struct_mon_nucl.label_seq_id _struct_mon_nucl.label_asym_id _struct_mon_nucl.label_alt_id _struct_mon_nucl.alpha _struct_mon_nucl.beta _struct_mon_nucl.gamma _struct_mon_nucl.delta _struct_mon_nucl.epsilon _struct_mon_nucl.zeta C 1 A . . . 29.9 131.9 222.1 174.2 G 2 A . 334.0 130.6 33.1 125.6 167.6 270.9 T 3 A . 258.2 178.7 101.0 114.6 216.6 259.3 # ---- abbreviated list -----