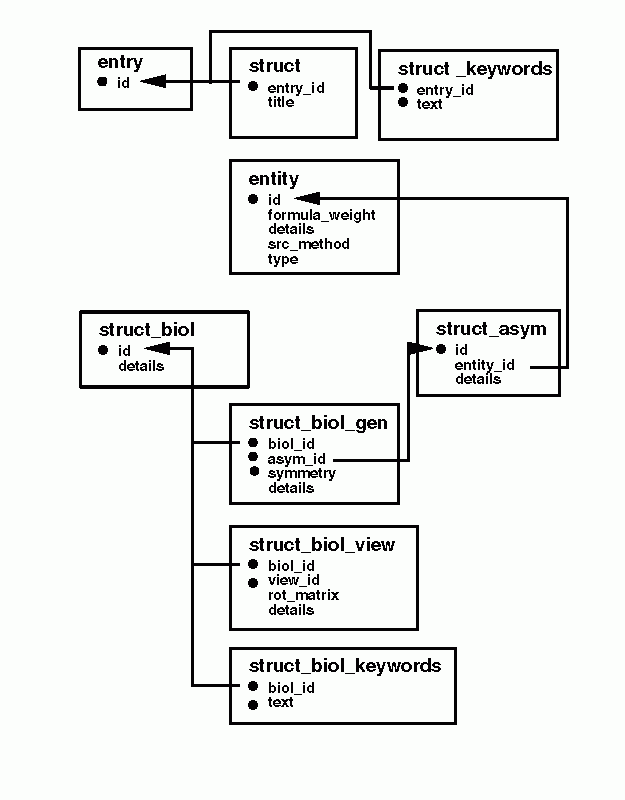
STRUCT category group. The top level description of the
structure within an mmCIF data file is contained in the
STRUCT category. The textual description of the structure
may include both a title and keywords. This description is labeled
with the top level identifier in an mmCIF data block, the _entry.id.
The crystallographic asymmetric unit
is described in the STRUCT_ASYM category. This category
lists the entities that were observed in the asymmetric unit and introduces
an identifier (_struct_asym.id) for each. This identifier
forms a part of the atom label in the ATOM_SITE category,
and this is used as an identifier in generating biological assemblies.
Biologically important assemblies are described in the collection of categories
descending from STRUCT_BIOL. Each biological assembly
is identified in the STRUCT_BIOL category and generated
from components of the asymmetric unit using crystallograhic symmetry
operations. Each assembly may by described by a keyword list and may
be presented in a number of different views.
The relationships between these categories in the STRUCT group
are shown in the following diagram.
In the following example for HIV protease (PDB 5HVP, Fitzgerald et al., 1990) there are two entities: the monomeric form of the enzyme and the drug inhibitor. The asymmetric unit contains two copies of each entity. Three biological assemblies are constructed for this system. One biological form contains the dimeric form of the enzyme and two others contain the dimeric enzyme plus one copy of the inhibitor.
# loop_ _struct_asym.id _struct_asym.entity_id _struct_asym.details A 1 'one monomer of the dimeric enzyme' B 1 'one monomer of the dimeric enzyme' C 2 'one partially occupied position for the inhibitor' D 2 'one partially occupied position for the inhibitor' # loop_ _struct_biol.id _struct_biol.details 1 ; significant deviations from twofold symmetry exist in this dimeric enzyme ; 2 ; The drug binds to this enzyme in two roughly twofold symmetric modes. Hence this biological unit (2) is roughly twofold symmetric to biological unit (3). Disorder in the protein chain indicated with alternative id 1 should be used with this biological unit. ; 3 ; The drug binds to this enzyme in two roughly twofold symmetric modes. Hence this biological unit (3) is roughly twofold symmetric to biological unit (2). Disorder in the protein chain indicated with alternative id 2 should be used with this biological unit. ; # loop_ _struct_biol_gen.biol_id _struct_biol_gen.asym_id _struct_biol_gen.symmetry 1 A 1_555 1 B 1_555 2 A 1_555 2 B 1_555 2 C 1_555 3 A 1_555 3 B 1_555 3 D 1_555
In the following example for the drug-DNA complex (NDB DDF040, Leonard et al., 1993) there are three entities in the asymmetric unit: one strand of DNA, drug and solvent. The biological assembly in this example is generated from the contents asymmetric unit and a second copy of the asymmetric unit transformed by the 8th symmetry operation of this space group.
_entry.id DDF040 loop_ _entity.id _entity.type _entity.src_method 1 polymer 'man' 2 non-polymer 'man' 3 water . _struct.entry_id DDF040 _struct.title ;5'-D(*TP*GP*GP*CP*CP*A)-3',ADRIAMYCIN ; _struct_keywords.entry_id DDF040 _struct_keywords.text ;DEOXYRIBONUCLEIC ACID ; loop_ _struct_asym.id _struct_asym.entity_id _struct_asym.details A 1 'NUCLEIC ACID' D 2 'ADRIAMYCIN' S 3 'H2O' _struct_biol.id 1 _struct_biol.details ;FULL DUPLEX DNA COMPLEXED WITH DRUG ; loop_ _struct_biol_gen.biol_id _struct_biol_gen.asym_id _struct_biol_gen.symmetry 1 A 1_555 1 A 8_555 1 D 1_555 1 S 1_555 1 D 8_555 1 S 8_555 loop_ _struct_biol_keywords.biol_id _struct_biol_keywords.text 1 'DNA' 1 'DNA DUPLEX' 1 'DNA DRUG COMPLEX'[an error occurred while processing this directive]