ATOM_SITE category. Atomic positions and their associated
uncertainties may be stored in either Cartesian or fractional coordinates,
and anisotropic temperature factors and occupancies may be stored for each position.
For each atomic position must be uniquely identified by the data item _atom_site.id.
Each position must also include a reference (_atom_site.type_symbol) to the table
of elemental symbols in category ATOM_TYPE. All other data items in
ATOM_SITE category are optional.
A typical atomic position for a macromolecule includes a variety of label information. The data items which label atomic positions can be divided into two groups: those that are integrated into higher level structural descriptions and those that are provided to hold a particular choice of nomenclature. These distinctions are described briefly in the following list of data items which may be used as part of an atom label.
_atom_site.label_alt_id
_atom_sites_alt.id defined in
category ATOM_SITES_ALT. This item is a unique
identifier for each alternative site in the ATOM_SITES_ALT list.
_atom_site.label_asym_id
_struct_asym.id defined in category
STRUCT_ASYM. This item identifies a particular entity in the
crystallographic asymmetric unit.
_atom_site.label_atom_id
_chem_comp_atom.atom_id defined in
category CHEM_COMP_ATOM. This identifier uniquely
identifies each atom within each chemical component.
_atom_site.label_comp_id
_chem_comp.id defined in category
CHEM_COMP. This item is the primary identifier for chemical components
which may either be mononers in a polymeric entity or complete non-polymer entities.
_atom_site.label_entity_id
_entity.id defined in the
ENTITY category. This item is used to identify chemically
distinct portions of the structure (e.g. polymer chains, ligands, solvent).
_atom_site.label_seq_id
_entity_poly_seq.num defined in the
ENTITY_POLY_SEQ category. This item is used to maintain the
correspondence between the chemical sequence of a polymeric entity and the
sequence information in the coordinate list and in may other structural categories.
This identifier has no meaning for non-polymer entities.
_atom_site.auth_asym_id
_atom_site.label_asym_id. This item is often used to hold
a strand or chain identifier.
_atom_site.auth_atom_id
_atom_site.label_atom_id. This item is often used to hold
a particular choice of atom names.
_atom_site.auth_comp_id
_atom_site.label_comp_id. This item is often used to hold
a particular choice of residue or ligand names.
_atom_site.auth_seq_id
_atom_site.label_seq_id. This item is often used to hold
a particular numbering scheme in the chemical sequence.
_atom_site.group_PDB
_atom_site.footnote_id
atom_sites_footnote.id
in category ATOMS_SITES_FOOTNOTE. This item is provided
as a means to include additional textual information about an atomic position
or about a group of atomic positions.
Alternative conformations are identified in the ATOM_SITE
category by the data item _atom_site.label_alt_id. This item
refers the parent item definition of the alternate site identifier in
category ATOM_SITES_ALT. The ATOM_SITES_ALT
category contains the list of each alternative conformation.
Ensembles of alternative conformations are identified in category
ATOM_SITES_ALT_ENS. Each ensemble is generated from one
or more of the alternate conformations listed in
in the list of alternative sites in category ATOM_SITES_ALT.
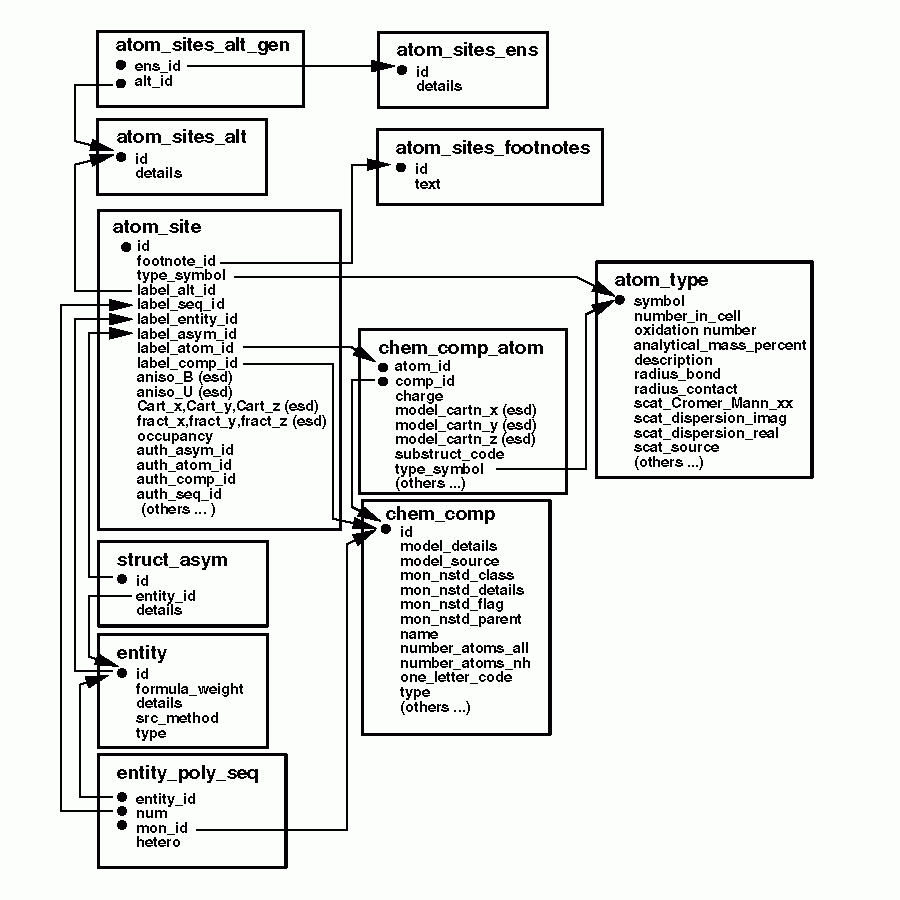
The relationships among this ATOM_SITE group of categories is
shown in the following figure.
An abbreviated example of the ATOM_SITE category.
loop_ _atom_site.group_PDB _atom_site.type_symbol _atom_site.label_atom_id _atom_site.label_comp_id _atom_site.label_asym_id _atom_site.label_seq_id _atom_site.label_alt_id _atom_site.Cartn_x _atom_site.Cartn_y _atom_site.Cartn_z _atom_site.occupancy _atom_site.B_iso_or_equiv _atom_site.footnote_id _atom_site.auth_seq_id _atom_site.id ATOM N N VAL A 11 . 25.369 30.691 11.795 1.00 17.93 . 11 1 ATOM C CA VAL A 11 . 25.970 31.965 12.332 1.00 17.75 . 11 2 ATOM C C VAL A 11 . 25.569 32.010 13.808 1.00 17.83 . 11 3 ATOM O O VAL A 11 . 24.735 31.190 14.167 1.00 17.53 . 11 4 ATOM C CB VAL A 11 . 25.379 33.146 11.540 1.00 17.66 . 11 5 ATOM C CG1 VAL A 11 . 25.584 33.034 10.030 1.00 18.86 . 11 6 ATOM C CG2 VAL A 11 . 23.933 33.309 11.872 1.00 17.12 . 11 7 ATOM N N THR A 12 . 26.095 32.930 14.590 1.00 18.97 4 12 8 ATOM C CA THR A 12 . 25.734 32.995 16.032 1.00 19.80 4 12 9 ATOM C C THR A 12 . 24.695 34.106 16.113 1.00 20.92 4 12 10 ATOM O O THR A 12 . 24.869 35.118 15.421 1.00 21.84 4 12 11 ATOM C CB THR A 12 . 26.911 33.346 17.018 1.00 20.51 4 12 12 ATOM O OG1 THR A 12 3 27.946 33.921 16.183 0.50 20.29 4 12 13 ATOM O OG1 THR A 12 4 27.769 32.142 17.103 0.50 20.59 4 12 14 ATOM C CG2 THR A 12 3 27.418 32.181 17.878 0.50 20.47 4 12 15 ATOM C CG2 THR A 12 4 26.489 33.778 18.426 0.50 20.00 4 12 16 ATOM N N ILE A 13 . 23.664 33.855 16.884 1.00 22.08 . 13 17 ATOM C CA ILE A 13 . 22.623 34.850 17.093 1.00 23.44 . 13 18 ATOM C C ILE A 13 . 22.657 35.113 18.610 1.00 25.77 . 13 19 ATOM O O ILE A 13 . 23.123 34.250 19.406 1.00 26.28 . 13 20 ATOM C CB ILE A 13 . 21.236 34.463 16.492 1.00 22.67 . 13 21 ATOM C CG1 ILE A 13 . 20.478 33.469 17.371 1.00 22.14 . 13 22 ATOM C CG2 ILE A 13 . 21.357 33.986 15.016 1.00 21.75 . 13 23 # - - - - Abbreviated - - - - HETATM C C1 APS C . 1 4.171 29.012 7.116 0.58 17.27 1 300 101 HETATM C C2 APS C . 1 4.949 27.758 6.793 0.58 16.95 1 300 102 HETATM O O3 APS C . 1 4.800 26.678 7.393 0.58 16.85 1 300 103 HETATM N N4 APS C . 1 5.930 27.841 5.869 0.58 16.43 1 300 104 # - - - - Abbreviated - - - -
An abbreviated example illustrating the description of alternate conformations an the HIV protease structure (PDB 5HVP, Fitzgerald et al., 1990).
loop_ _atom_sites_alt.id _atom_sites_alt.details '.' ; Atom sites with the alternative id set to null are not modeled in alternative conformations ; '1' ; Atom sites with the alternative id set to 1 have been modeled in alternative conformations with respect to atom sites marked with alternative id 2. The conformations of amino acid side chains and solvent atoms with alternative id set to 1 correlate with the conformation of the inhibitor marked with alternative id 1. They have been given an occupancy of 0.58 to match the occupancy assigned to the inhibitor. ; '2' ; Atom sites with the alternative id set to 2 have been modeled in alternative conformations with respect to atom sites marked with alternative id 1. The conformations of amino acid side chains and solvent atoms with alternative id set to 2 correlate with the conformation of the inhibitor marked with alternative id 2. They have been given an occupancy of 0.42 to match the occupancy assigned to the inhibitor. ; '3' ; Atom sites with the alternative id set to 3 have been modeled in alternative conformations with respect to atoms marked with alternative id 4. The conformations of amino acid side chains and solvent atoms with alternative id set to 3 do not correlate with the conformation of the inhibitor. These atom sites have arbitrarily been given an occupancy of 0.50. ; loop_ _atom_sites_alt_ens.id _atom_sites_alt_ens.details 'Ensemble 1-A' ; The inhibitor binds to the enzyme in two, roughly twofold symmetric, alternative conformations. This conformational ensemble includes the more populated conformation of the inhibitor (id=1) and the amino acid side chains and solvent structure that correlate with this inhibitor conformation. Also included are one set (id=3) of side chains with alternative conformations when the conformations are not correlated with the inhibitor conformation. ; loop_ _atom_sites_alt_gen.ens_id _atom_sites_alt_gen.alt_id 'Ensemble 1-A' '.' 'Ensemble 1-A' '1' 'Ensemble 1-A' '3'